Investigation of Innate Immune Responses against HIV with Camelid Nanobodies

Project Member

Prof. Dr. Eicke Latz
Project Leader
Prof. Dr. Eicke Latz
Phone: +49 228 287 – 51239
Fax: +49 228 287 – 51221
Eicke.Latz@uni-bonn.de
Institut of Innate Immunity
Biomedical Center, 1G007
University Hospital Bonn
Venusberg-Campus 1
53127 Bonn
Germany

Dr. Florian I. Schmidt
Institut of Innate Immunity
Biomedical Center, 1G011
University Hospital Bonn
Venusberg-Campus 1
53127 Bonn
Germany

Jennifer Würth,
Postdoc
Project Summary
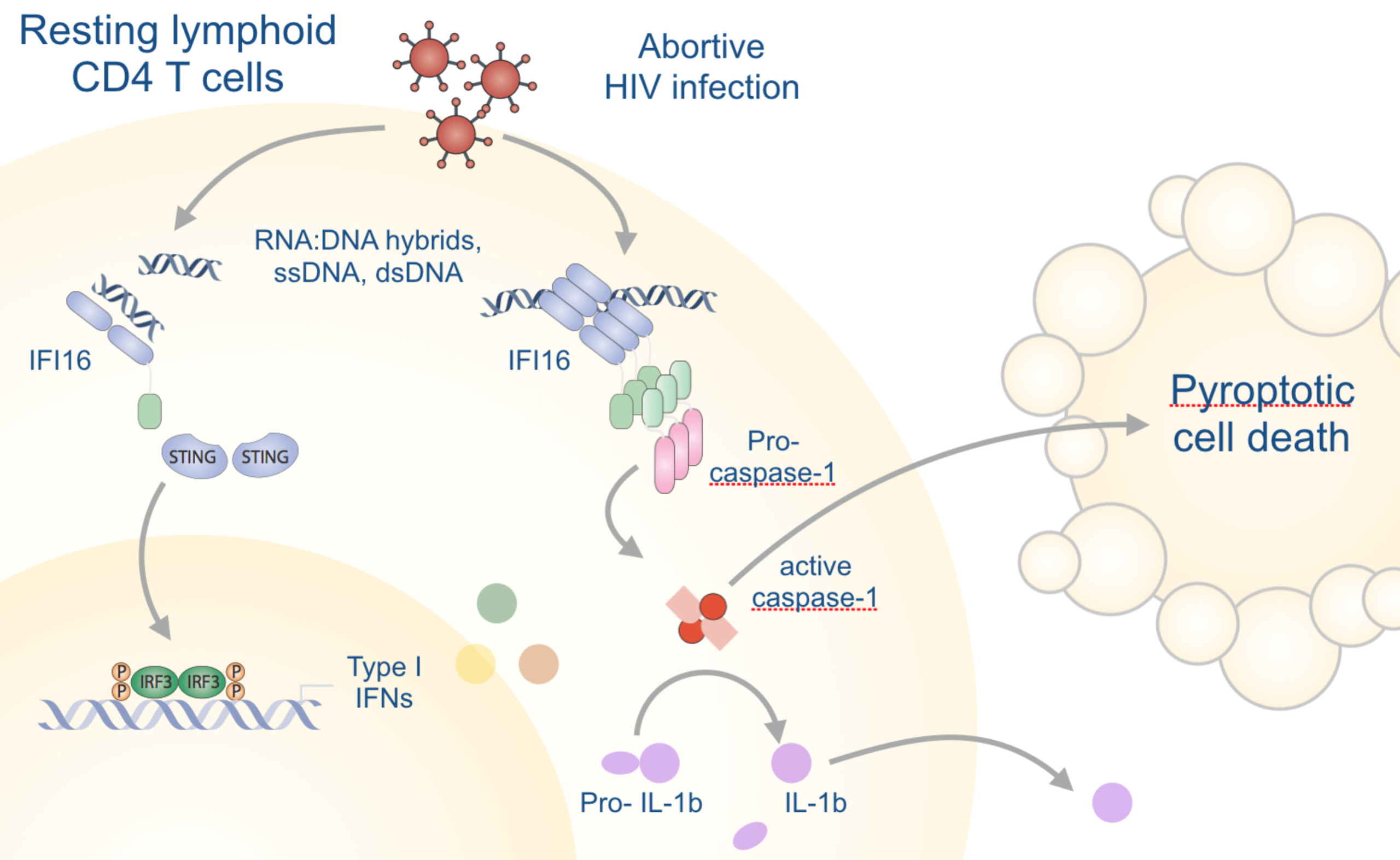
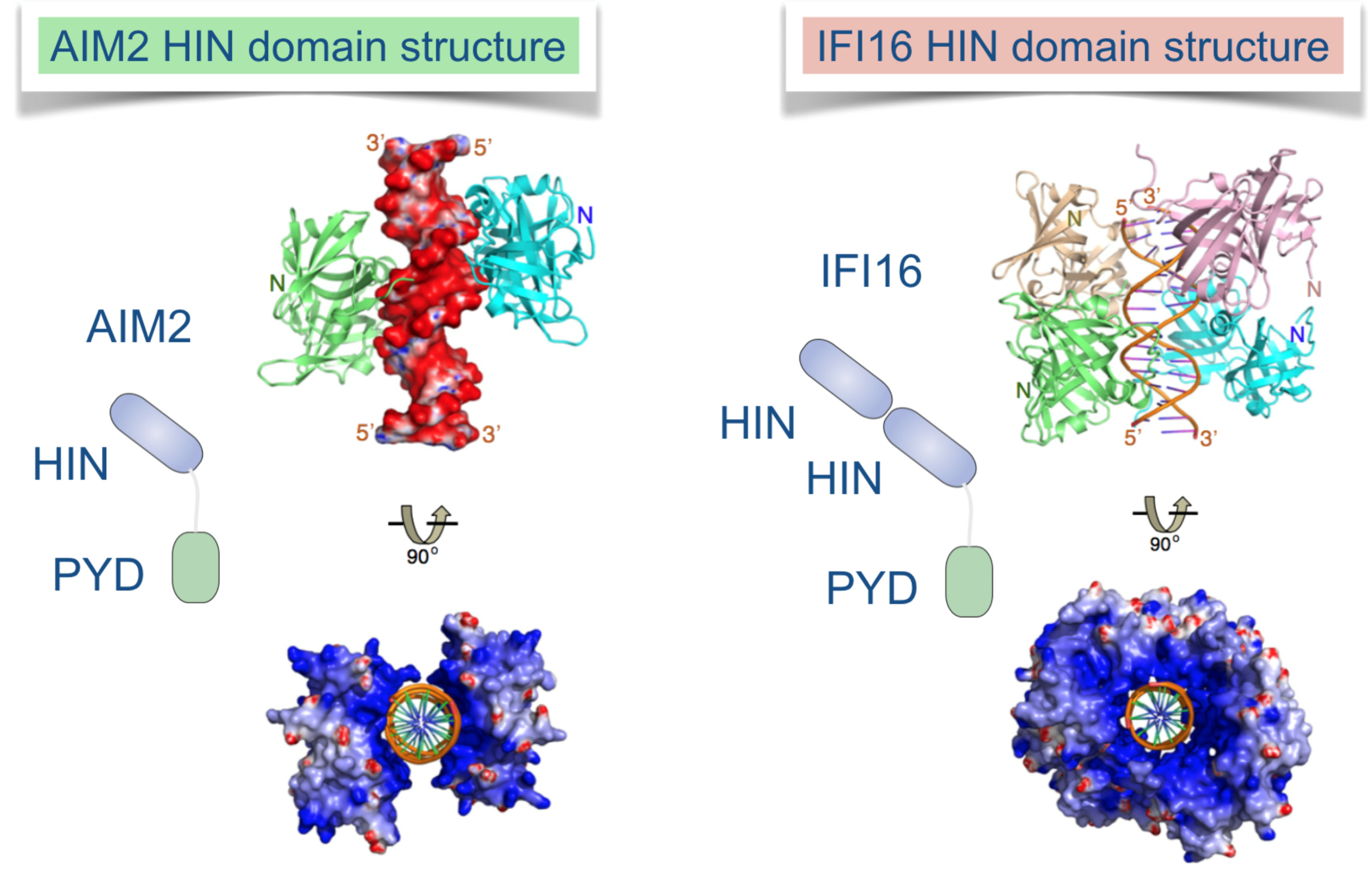
Germ line-encoded receptors and adaptors sense signs of infection and mount the appropriate innate immune response. While these responses can curtail infection with human immunedeficiency virus (HIV), aberrant activation of inflammation is also linked to HIV pathogenesis. Although the massive loss of CD4 T cells in the course of disease progression has been linked to pyroptosis, molecular details of HIV-induced inflammasome assembly and other fundamental aspects of the innate immune response to HIV are not understood.
While previous studies have mostly relied on cytokine measurements and indirect assessment of cell loss, we propose a different approach to investigate innate immune responses to HIV infection:
We aim to directly visualize and quantify inflammasome assembly and antiviral responses in human primary cell (mixtures) with novel inflammasome biosensors and nanobodies. Nanobodies are single domain antibodies derived from camelid heavy chain-only antibodies, which we will custom generate and equip with additional functionalities by enzymatic or genetic modification. We will detect individual cells that assemble inflammasomes or mount antiviral transcriptional responses by flow cytometry and ImageStream analysis. HIV strains will be equipped with a novel genetically-encoded inflammasome biosensor that can reveal inflammasome assembly in productively or abortively infected primary cell cultures, including PBMCs and tonsillar human lymphocyte aggregate cultures (HLAC). This will allow us to sort responding cells and analyze their transcriptome. Nanobodies will be used to visualize the involved molecular interactions by advanced microscopy techniques, identify novel interactions, and deplete proteins in primary cells for functional analysis.
We will further dissect the molecular regulation of NLRX1, a critical anti-inflammatory host factor required for HIV infection in myeloid cells that is upregulated early in SIV-infected macaques. Using custom-generated nanobodies against NLRX1, we will evaluate its function in HIV-infected primary cells, analyze its regulation by oligomerization, post-translational modification, or localization, and will identify novel interaction partners by proximity labeling approaches. We postulate that NLRX1 is an intricately regulated signaling hub, which integrates input to broadly coordinate anti-inflammatory processes.
We will generate, apply, and share novel tools to visualize inflammatory responses and their regulation. Our work will thus help reveal how HIV infection triggers and avoids the innate immune system in relevant primary cell models.