Investigation of cellular mechanisms of PQBP1-dependant innate signaling of HIV‑1

Project Member

Dr. Renate König
Project Leader
Dr. Renate König
Phone: +49 6103 77 – 4019
Fax: +49 6103 77 – 1255
Renate.Koenig@pei.de
RKoenig@sbpdiscovery.org
Paul-Ehrlich-Institut
Federal Institute for Vaccines
and Biomedicines
Paul-Ehrlich-Str. 51 – 59
63225 Langen
Germany
Immunity and Pathogenesis Program
Sanford Burnham Prebys (SBP)
Medical Discovery Institute
La Jolla, CA 92037, USA

Dr. Nina Hein-Fuchs,
Postdoc

Maïwenn Bergez,
PhD Student
Project Summary
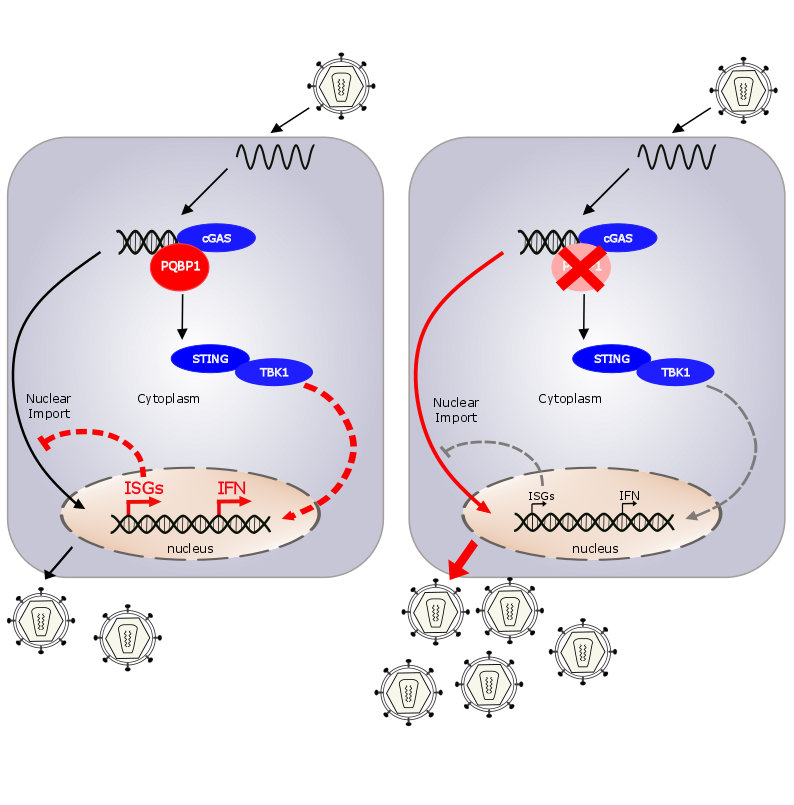
In a collaborative publication, PQBP1 was identified as an essential component of the cGAS/IRF3-dependent innate response to HIV. PQBP1 binds to HIV1 DNA after reverse transcription, and activates cGAS to produce the second messenger cGAMP to induce STING/IRF3 signalling. Interestingly, the requirement for PQBP1 appears to be specific for lentiviral DNA, as other types of DNA, including poxvirus, did not require PQBP1 to stimulate innate immune responses. However, specific details of the PQBP1/cGAS innate receptor complex and the underlying mechanism influencing sensing are not known (Yin et al., 2020). Therefore, in the first funding period, we were interested, whether other members of the Retroviridae family are dependent on cGAS/IRF3- and PQBP1-dependent innate responses (Bergez et al., 2019). Furthermore, we generated and characterized hiPSCs from PQBP1-mutated Renpenning-syndrome patients and healthy donor controls in order to provide an unlimited pool of relevant primary innate cells to analyze PQBP1 requirement and functions (Fuchs NV et al., 2020; Fuchs NV et al., 2019). The main goals of the new funding period are to define the specificity of PQBP1-dependent sensing and the underlying mechanisms. Initially, we will examine other genera of the Retroviridae family in comparison to DNA viruses to understand the breadth of PQBP1 recognition. Moreover, we aim to detect the reverse transcribed DNA intermediates bound to PQBP1 in order to understand their commonalities. We aim to analyze whether cGAS is required for this step. Furthermore, we are interested in the underlying mechanisms. We will determine whether infection of viruses recognized by PQBP1 specifically enhance the binding between PQBP1 and cGAS and whether specific PTMs are involved. Our results will provide new insights into the cellular mechanism underlying PQBP1-dependant innate signaling of HIV‑1.