SPP 1923:
“Innate Sensing and Restriction of Retroviruses”
DFG Priority Program.
Retroviruses comprise a diverse group of exogenous and endogenous viruses defined by their unique replication strategy to reverse-transcribe their RNA genome into a complementary DNA. Millions of years of coevolution with their mammalian hosts gave rise to highly pathogenic as well as apathogenic members of this family of viruses. Cell-type specific cell-autonomous components of the innate immune system, including specialized pattern recognition receptors (PRRs) and broadly antiviral restriction factors (RFs), represent key determinants of the fundamentally different outcomes of retroviral infections. Specific parameters under consideration include the efficiency of cross- and intra-species transmission, pathogenesis, virus evolution, and the ability to establish a chronic infection in a new host.
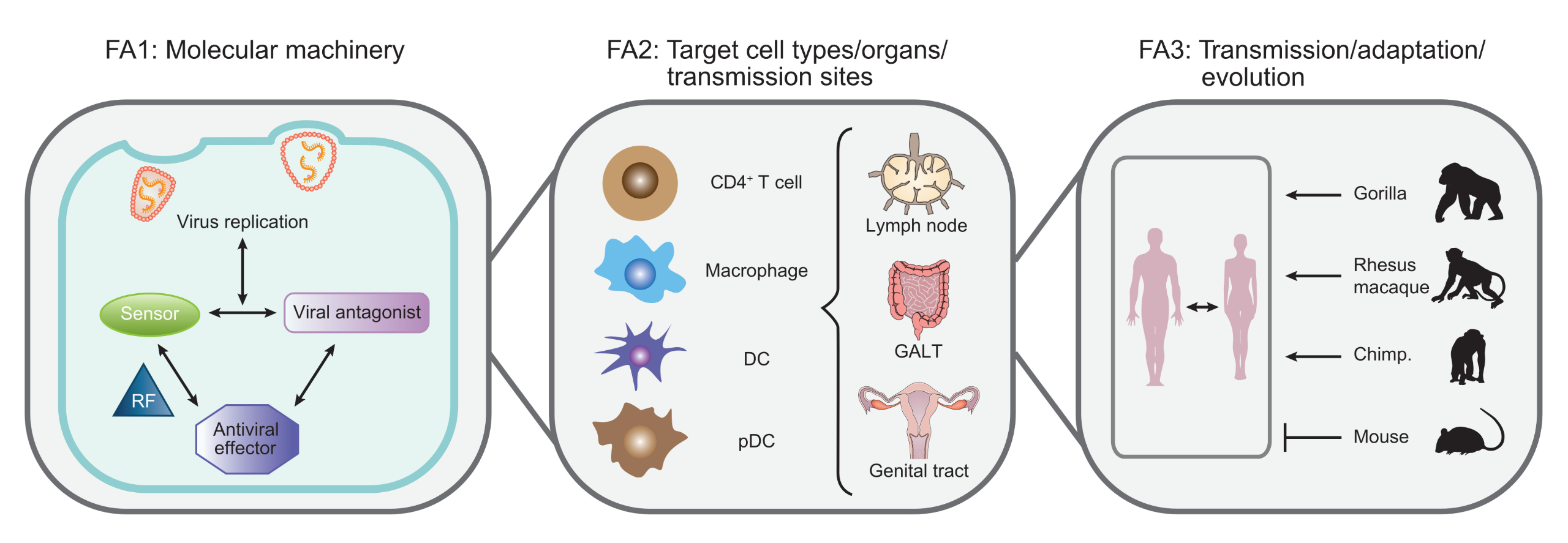
The identification of a series of PRRs and RFs that contribute to cell-autonomous immunity against retroviruses provided a strong impetus for the establishment of the interdisciplinary national research network SPP1923. The overall goal of SPP1923 is to integrate experts in retrovirology and innate immunity to assemble a national network that addresses key open questions on the interplay of retroviruses with the cell-autonomous immune system of their hosts in collaborative projects. By these combined efforts, we aim at the identification of the full molecular sensing and restriction machinery involved, its regulation, evolved virus-encoded countermeasures, and pathophysiological consequences. Pathogenic and apathogenic retroviruses will be investigated in cell systems ranging from monotypic cell cultures to complex ex vivo and animal models (Fig. 1). At these different levels of complexity, SPP1923 aims at dissecting mechanisms of innate immune recognition, viral evasion and counteraction thereof and at defining the functional consequences of these virus-host interactions.
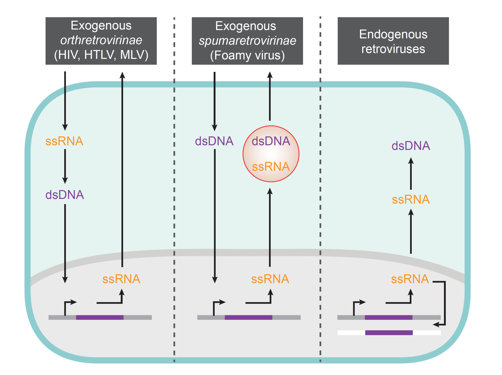
Within the family of retroviruses, several variations of this replication strategy exist that result in exposure of distinct PAMPs to host cell PRRs at different stages of their life cycle (Fig. 2). Elucidating the similarities and differences of the interplay and pathological impact of these different classes of retroviruses with the innate immune system of humans, non-human primates and rodents will provide insights into both virus and host coevolution and constitutes another central goal of this initiative. Classes of retroviruses studied within SPP1923 include pathogenic exogenous orthoretroviruses (e.g. HIV‑1), Spumaretroviruses (e.g. Foamy virus) as well as endogenous retroviruses and retroviral elements.
Fig. 1: Topology of the addressed field of research at the interface of retrovirus biology and cell-autonomous immunity. Work in classical cell culture systems (left) seeks to identify and understand at the molecular level the full set of cell-autonomous recognition factors of retroviruses and resulting effector functions at high spatio-temporal resolution. These events underlie cell system-specific regulation and their relevance is evaluated in primary cell and organotypic infection models (middle). The impact of innate recognition is defined for retroviral cross-species and human-to-human transmission, evolutionary virus-host adaptation including polymorphisms in innate genes/viral antagonists, and pathogenesis in the new host (right).
Fig. 2: Schematic of retroviral replication strategies and genomic nucleic acid species involved.
Funded by: